"Enabling care-givers in their medical decision making process to apply the right and only right therapy or medication, based on the unique patient genetic profile. This, by making this technology available in a decentralized way: more affordable, faster, and more accessible to hospitals and patients, without complex workflows or heavy data analysis. This would represent a major step forward in personalized oncology care."
Our technology: PAP-PCR: Nucleic Acid Amplification and Detection
PAP-PCR: a PCR platform based on the Prephosphorylation Activated Polymerisation step in a PCR reaction.
The technology is a new nucleic acid amplification technology that has surprising properties for nucleic acid amplification (Liu and Sommer, 2004). For example, its amplification selectivity, or signal to noise ratio, is so extremely high that it can detect a single copy of DNA mutant molecule in 1 billion of almost identical wild type molecules. This level of selectivity is over 1,000,000 times more than that of PCR or any other technologies. In addition, its sensitivity or the detectable smallest copy number of the target molecule can reliably arrive at a single copy level. This level of sensitivity is over 100 times more than that of PCR technology.
PAP-PCR enables accurate low-frequency variant detection (VAF 0,001% for single target detection. Ranging from 0,2 to 1 ppm for multiple targets) across multiple platforms, including qPCR, and next generation sequencing.
Our technology combines conventional quantitative PCR (qPCR), a technology that quantifies amplified targeted DNA sequences, with our unique PAP thermophilic enzymes and blocked primers carrying a special 3’ terminal modification that is removed only when the primer perfectly matches the target mutation, preventing non-specific extension. By integrating this design with qPCR, we eliminate nearly all false positives and exceed conventional PCR detection limits by a factor of 10E6. Also detection of DNA molecules containing mutations and wild-type background molecules, by using NGS platforms is demonstrated.
With this technique, a single copy of a mutation can be detected in a background of as many as one billion wild-type plasmid copies, or in 100,000 copies of genomic DNA (gDNA). In comparison, with conventional PCR, false positive signals are already generated from about 100 wild type copies. By using NGS as a “detector” we were able to detect a few copies of mutant in a background of 100,000 wild-type molecules.
This makes our technology extremely suitable for cfDNA mutation analysis, early cancer diagnostics and monitoring disease progression.
Key Characteristics
Selectivity
One in a billion (1:10E9): unmatched detection of ultra-low-abundance mutations.
Ultra-early detection
Catching cancer at stage 0.
Cost and turnaround time
Less complex workflow vs NGS, enabling faster results.
Scalable platform
Adaptable to multiple cancer types, facilitating pipeline growth.
Scalable technology
Adaptable to multiple cancers: ensuring broad market penetration.
Patient-friendly and reliable
Non-invasive testing with high sensitivity and speed.
How it works
Based on a method in which a unique and patented DNA polymerase serially couples pyrophosphorolysis and polymerization.
- 1st: the polymerase removes the blocked 3'-nucleotide ( T* ) from primer P*
- 2nd: it catalyzes polymerization: elongation of the primer (in the presence of dNTPs, without the blocked 3'-nucleotide) to form a polynucleotide product.
A blocked primer P* and a template are shown. When the blocked primer matches the template, polymerase catalyses pyrophosphorolysis and polymerization in the given order.
During the thermal cycling steps, amplification of the polynucleotide product results in the amplicon containing the mutation, ONLY. Visualisation is by intercalating dyes, NGS or TaqMan bases probes.
The future of cancer care lies in prevention rather than treatment. Genetic Tools develops breakthrough genetic detection technologies that detect cancer at an ultra-early stage—long before disease symptoms appear OR re-appear.
The first clinical tests for some of the most common biomarkers will be carried out together with several Academic Centers in the Netherlands. Test data will of course be published.
With test methods that are 1 million times more specific and 1,000 times more sensitive than current techniques, Genetic Tools makes cancer care proactive, personalized and preventive.
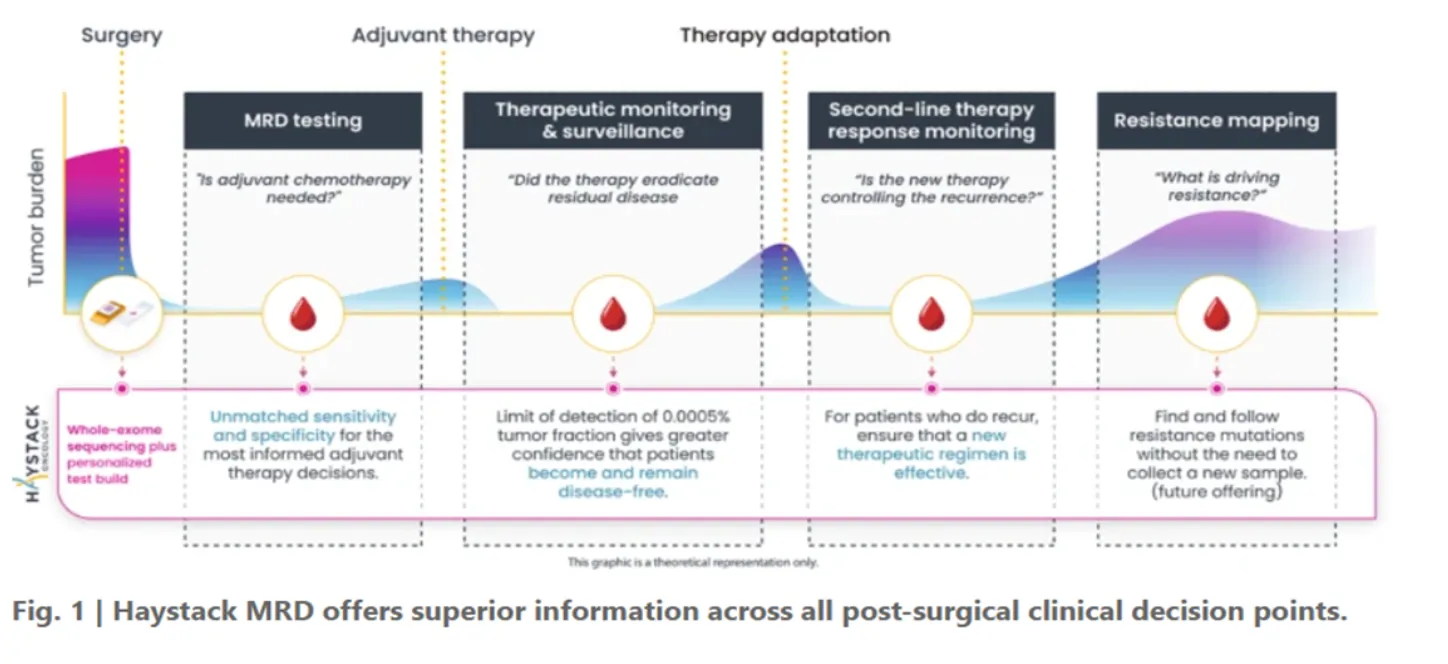
Utility in clinical decision making
Our MRD testing offers superior information across all post-surgical clinical decision points: guiding adjuvant therapy, monitoring residual disease and therapy response, detecting recurrence early, and mapping resistance drivers.
The future of cancer care lies in prevention, rather than treatment.
Genetic Tools develops breakthrough genetic detection technologies that detect cancer at an ultra-early stage, long before disease symptoms appear OR re-appear.
The first clinical tests for some of the most common biomarkers will be carried out together with several Academic Centers in the Netherlands. Test data will of course be published.
With test methods that are 1 million times more specific and 100 times more sensitive than current techniques, Genetic Tools makes cancer care proactive, personalized and preventive.
Key Differences
To understand effective cancer research the sensitivity, specificity and variant allele frequency of a testing methodology need to be understood. Herewith some explanations:
| Parameter | Definition | Importance |
|---|---|---|
| VAF | The fraction of DNA molecules carrying a mutation. | Helps determine tumor burden and detect emerging mutations. |
| Specificity | Ability to avoid false positives (detect only real mutations). | Ensures accuracy and prevents misdiagnosis. |
| Sensitivity | Ability to detect true positives (avoid missing mutations). | Important for detecting mutations at low levels and guiding treatment. |
Liquid Biopsy for Minimal Residual Disease (MRD) Monitoring
A patient with colorectal cancer undergoes surgery to remove the tumor. After surgery, a liquid biopsy (circulating tumor DNA, ctDNA) is performed to check for minimal residual disease (MRD).
- High Sensitivity is Critical The test must detect very low levels of ctDNA (e.g., VAF < 0.1%) to catch early signs of recurrence.
- High Specificity Prevents False Positives If the test wrongly detects mutations that aren't present, the patient might receive unnecessary chemotherapy.
- Challenge: If the sensitivity is too low, small traces of tumor DNA might be missed (false negative). If specificity is too low, normal DNA variations might be misinterpreted as cancerous mutations. (false positives)
Companion Diagnostics for Targeted Therapy (EGFR Mutations in Lung Cancer)
A patient with non-small cell lung cancer (NSCLC) is tested for EGFR mutations to determine eligibility for targeted therapy (e.g., Osimertinib).
- High Sensitivity is Required Some EGFR mutations (e.g., T790M, which causes resistance to first-line therapy) may be present at low VAF (1-5%) in tumors. Missing them could lead to ineffective treatment choices.
- High Specificity Prevents Misdiagnosis False positives could lead to giving the wrong drug, causing unnecessary side effects.
Clonal Hematopoiesis in Liquid Biopsies (False Positives Due to Aging)
A patient undergoing ctDNA testing for lung cancer has a detected TP53 mutation at VAF ~2%. However, they do not have a tumor with this mutation.
- Issue The detected mutation is from clonal hematopoiesis of indeterminate potential (CHIP)—a normal aging-related process where blood cells acquire mutations.
- Impact on Specificity If the test lacks specificity, CHIP mutations could be misclassified as tumor-derived, leading to unnecessary treatment.
- Solution: Some assays incorporate white blood cell sequencing to distinguish CHIP mutations from true tumor-derived mutations.
Contact Details
- info@genetictools.eu
- General inquiries: Ronald at +31 621929402
- Technical assistance: Anton at +31 6 245 626 28